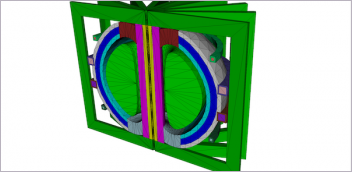
The compound in this image (in gray) was calculated to bind to the SARS-CoV-2 spike protein (cyan) to prevent it from docking to the human ACE2 receptor (in purple). Image courtesy of Micholas Smith/ORNL, U.S. Department of Energy.
Latest News
April 10, 2020
The COVID-19 pandemic, which swept through China and Europe, and is rapidly spreading throughout the United States, has upended nearly every facet of life. With schools and most businesses closed, people are adjusting to sheltering in place. Meanwhile, scientists are scrambling to create new ways to diagnose and treat this new virus while healthcare providers struggle to acquire vital equipment like protective masks, face shields, and ventilators.
The technology industry—and specifically, many in the engineering community—have risen to the occasion. Around the world, technology providers, universities, private industry, and individual engineers and hobbyists have joined forces to address both these supply chain and research challenges.
Supercomputers Target COVID-19
Powerful compute resources are being deployed to help improve diagnoses and identify new treatments for COVID-19. At Oak Ridge National Laboratory (ORNL) in Tennessee, researchers used the Summit supercomputer to identify 77 small-molecule drug compounds that could potentially bind to the glycosylated spike (S) protein, through which the COVID-19 virus accesses host cells. One or more of those compounds could potentially disarm the virus and point the way to future treatments.
According to the South China Morning Post, researchers in China are also leveraging the Tianhe-1 supercomputer and artificial intelligence (AI) to diagnose COVID-19 patients using chest scans. The National Supercomputer Centre in Tianjin claims the system was able to provide a diagnosis wihtin 10 seconds based on CT scan images.
The i2b2 tranSMART Foundation and its members are leveraging the Foundation’s open source platforms to rapidly create a federation of translational research centers. This federation will enable large-scale population monitoring of COVID-19 patients, mobilizing the data from their network of more than 200 institutions worldwide to identify and collect datasets and tools that can be leveraged to study COVID-19 by the scientific and clinical research community. This timely, comprehensive data on COVID-19 patients across the globe is helping to broker research on prevention and cures, including identifying “hot spots” for where medical resources are most needed.
Organizations interested in supporting these efforts can find more information here.
Searching for Treatments
At ORNL researchers built a model of the coronavirus spike protein based on the earlier studies of the structure. The Summit computer used NVIDIA V100 GPUs and GROMACS, a GPU-accelerated simulation package for biomolecular systems, to run the team’s experiments on more than 8,000 compounds. The results were published last month on ChemRxiv, a preprint server for chemistry research.
“We were able to design a thorough computational model based on information that has only recently been published in the literature on this virus,” said team member and UT/ORNL CMB postdoctoral researcher Micholas Smith. Smith referred to a previous study published in Science China Life Sciences that modeled the spike protein for risk of human transmission. When Chinese researchers sequenced the virus, they found that it infects the body via a mechanism similar to that of the SARS virus.
The power of the Summit supercomputer made it possible to rapidly evaluate a large number of potential compounds. The team identified 77 small-molecule compounds, including medications and natural compounds, that could be candidates for further experimental testing. In the simulations, the compounds bind to regions of the spike that are important for entry into the human cell.
Researchers at the University of Texas at Austin have since released a more accurate model of the S protein, and the ORNL researchers plan to use that model to re-run the study.
IBM, which built the Summit, is also providing other resources to help researchers address the pandemic. According to IBM, the company’s Watson Health unit is working directly with health organizations around the world to better understand the nature of COVID-19.
Advanced computing clusters from Dell are being used to help better understand disease outbreaks, as well as how to track their spread. For example, The University of Texas at Austin and other institutions utilized the Texas Advanced Computing Center’s (TACC) Wrangler system to analyze comprehensive travel data from location-based services to develop a model of the spread of the virus through China.
Technology Rx
Companies like Dell, NVIDIA, and others are providing financial support for COVID-19 efforts, but are also donating technology to key organizations, including laptops, workstations, and compute resources. In China, Dell provided an IT infrastructure donation to the Hubei Center for Disease Control and Prevention (CDC) which allowed the Center to respond more quickly to the epidemic.
NVIDIA is giving away free 90-day licenses of its genome-sequencing software Parabricks to researchers in the fight against Coronavirus. According to the company's blog post, “In response to the current pandemic, NVIDIA is sharing tools with researchers that can accelerate their race to understand the novel coronavirus and help inform a response.”
Oracle Cloud Infrastructure has released an updated NVIDIA GPU Cloud Machine Image in support of these efforts. Because the software takes advantage of the computing cores in the GPU, “data that once took days to analyze can now be done in under an hour,” NVIDIA says.
To gain access, researchers need to fill out a request form.
NVIDIA has also joined the COVID-19 High Performance Computing Consortium, which has brought together leaders from the U.S. government, industry and academia to accelerate COVID-19 research using HPC resources.
Folding@home
Engineers and gamers with workstations in their home can also help. Folding@home, a distributed computing project to research protein dynamics, is now devoting its resources to simulate the dynamics of COVID-19.

On March 15, Greg Bowen, the project's co-director, wrote, “We’re simulating the dynamics of COVID-19 proteins to hunt for new therapeutic opportunities ... Downloading Folding@home and helping us run simulations is the primary way to contribute.”
For those that participate in the project, their unused computing cycles during idle times become part of the distributed computing infrastructure to simulate and render COVID-19's behavior.
“While you keep going with your everyday activities, your computer will be working to help us find cures for diseases like cancer, ALS, Parkinson’s, Huntington’s, Influenza and many others,” Folding@home explains.
“GPUs have the possibility to perform an enormous number of Floating Point Operations (FLOPs) ... After much work, we have been able to write a highly optimized molecular dynamics code for GPU’s, achieving a 20x to 40x speed increase over comparable CPU code for certain types of calculations,” wrote Folding@home.
Major workstation and GPU providers are supporting these efforts through their own user communities. To download the app and join the project, go to this link.
Subscribe to our FREE magazine, FREE email newsletters or both!
Latest News